Medications
Systemic lupus erythematosus (SLE)
We used Vanderbilt’s Synthetic Derivative (SD), a de-identified version of the EHR, with 2.5 million subjects. We selected all individuals with at least one SLE ICD-9 code (710.0) yielding 5959 individuals. To create a training set, 200 were randomly selected for chart review. A subject was defined as a case if diagnosed with SLE by a rheumatologist, nephrologist, or dermatologist.
Type 1 and type 2 Diabetes Mellitus
This document describes the Stanford University algorithm to extract individuals with diabetes and the type of diabetes from electronic health records (EHRs). There are two main tasks of this phenotype development: 1) to extract patients with diabetes (gestational diabetes is excluded), and 2) to discriminate between type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM). Instead of identifying all diabetes cases, we aim to reduce the number of false positives in our diabetes cohort.
Type 1 Diabetes
Phenotype Algorithm for Type 1 Diabetes – eMERGE Phase-IV Program
Type 2 Diabetes - Demonstration Project
Type 2 Diabetes phenotype algorithm for the DNA Databank Demonstration Project.
Type 2 Diabetes - PRS Evaluation
NOTE:
The following files were updated on 4/9/2021 so that the output of the #feature table in the eMERGE_IV_OMOP_T2DM_PRS_algorithm script matches the data dictionary.
Files:
- T2DM_DD_Feature_Count_OMOP_2021049.csv
- eMERGE_IV_OMOP_T2DM_PRS_algorithm_20210409.txt
- eMERGE_IV_OMOP_T2DM_PRS_algorithm_20210409.zip
Type 2 Diabetes Mellitus
Type1 or Type 2 Diabetes Mellitus
Phenotyping algorithm for the identification of patients with type 1 or type 2 diabetes mellitus (DM) preoperatively using routinely available clinical data from electronic
health records.
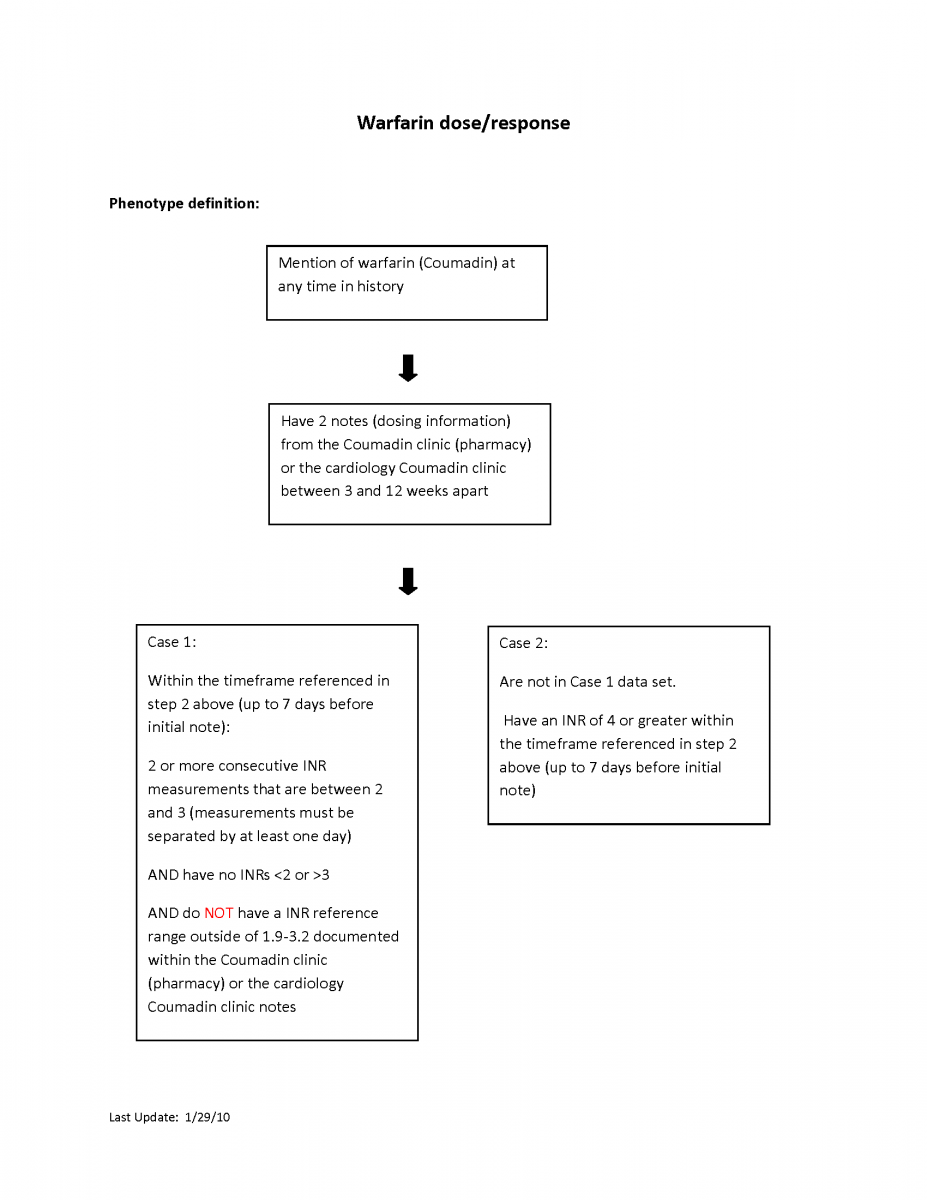
Warfarin dose/response
This algorithm identifies patients who have a stable within-range INR (assuming a target INR of 2-3) over at least a three week period and correlates with their warfarin weekly dose. It is used to identify pharmacogenetics behind warfarin stable dose.
White Blood Cell Indices
Genetic variation that predicts white blood count (WBC) and it differential, a marker of the health of the immune system.
WBC is unique among the identified inflammatory predictors of chronic disease in that it has been routinely measured in healthy patients in an unbiased way for the duration of the electronic medical record data.
